Howdy!!!
The NDEx Team would like to share some news and hopes you'll find it interesting!
Follow us on LinkedIn or visit our website to stay informed on new project features, releases and other interesting content and subscribe to our YouTube channel!
Questions? Comments? We ‘d be happy to hear from you!
NDEx v1.2 Release Candidate 1 available now!!!
The NDex Team is proud to announce that the NDEx v1.2 Release Candidate is now available on our Test and Public Servers… We will keep testing, optimizing and minor changes will be applied in the next few days; the official release date for NDEx v1.2 is scheduled for June 17th and we are right on track!
Biologists, bionformatic researchers and developers, head over to our website and START using NDEx NOW!
NDEx v1.2 is rich in features and improvements including a new display table for nodes/edges properties, a redesigned provenance history, improved error logging, a new advanced query option, full BioPAX3 support and a new clean, modern look. We also released a brand new copier program (ndex-sync v1.2) that can now handle network updates. Check out our website in the next couple of weeks for updated documentation, new tutorials, release notes and don't forget to keep an eye on our blog!
Biological networks and infectious diseases
This month we feature a mini-review by Mulder and collaborators that focuses on the advantages of using biological network models to improve our understanding of infectious diseases. Specifically, Mulder and collaborators discuss the use of biological networks for function prediction, comparison in evolutionary studies as well as generation and use of host-pathogen interaction networks, all applied to the infectious disease Tuberculosis, its causing agent, Mycobacterium tuberculosis and its favorite host… Homo sapiens!
READ FULL ARTICLE IN PMC >>
New collaborations for NDEx
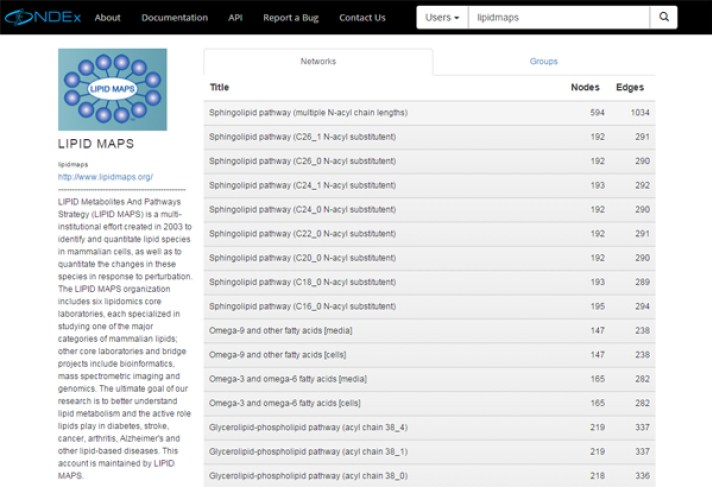
The NDEx Project has successfully started new collaborations with the "LIPID Metabolites And Pathways Strategy (LIPID MAPS)" and the "Binding Database" to provide easy access to new valuable data on lipid metabolites and small molecules binding measurements.
The lipid metabolites networks (BioPAX3 format) are available on the NDEx Public Server and maintained by the LIPID MAPS team: click the image below to browse the LIPID MAPS networks on NDEx!
Ligand-target binding measurement data from the Binding Database will be available later this summer.